A short keynote review of Course at school: Introduction to Bioinformatics (1001)
声明
一些我很熟悉的算法会略过过程,不表示不是重点
Chapter 1 Introduction to bioinformatics
Definition of Bioinformatics
Bioinformatics: an interdisciplinary field that develops and applies computer and computational technologies to study biomedical questions
The Bio- in Bioinformatics
From Genotype to Phenotype:
- Genome/DNA
- RNA
- Proteins
- Molecular Networks(分子网络)
- Cells
- Physiology/Disease
The -informatics in Bioinformatics
- Management
- Computation
- Mining
- Modeling/Simulation
Chapter 2 Gene and RNA
What is Gene?
A gene is a locus (or region) of DNA that encodes a functional protein or RNA product, and is the molecular unit of heredity.
The central dogma and splicing
DNA -> (transcription) -> Pre-mRNA -> (splicing) -> mRNA -> (translation) -> protein
Gene Finding Approach
Computational Methods:
- Something that matches statistical patterns common to all qenes (ab initio)
- Something that matches an already known gene (homology)
- Hybrid
Things Measured About Genes
- ORF
- Codon Usage -> CAI
- Features and motifs
- Promoters, splice sites, enhancers, untranslated regions (UTRs)
Similarity-based Approach to Gene Prediction
Given a known gene and an un-annotated genome sequence, find a set of substrings of the genomic sequence whose concatenation best fits the known gene
EXON CHAINING PROBLEM
DP
Non-coding RNA(ncRNA)
RNA molecules that function without being translated into a protein
non-coding RNAs is not always non-translatable
- tRNAs
- miRNA
SNP
Single Nucleotide Polymorphism— Mutation of a single nucleotide (A,C,T,G)
The most simple form and most common source of genetic polymorphism in the human genome
CNV
Copy number variation(CNV) of DNA sequences constitute arge segments of DNA ranging trom 1 Kb that have copy number ditterences when compared to a reterence genome
CNVs:
- deletions
- duplications
- insertions
CNVs may influence gene expression and adaptation, can give insight into complexity of normal phenotypic variation and disease.
Genome Wide Association Study
GWAS is the study of genetic variation across the entire genome that is designed to associate genetic variations (SNPs) with traits or with the presence or absence of disease or condition.
Single cell RNA sequencing
- Dissociation
- Isolation
- Single
- RNA extraction
- cDNA senthesis
- Single-cell sequencing
- Expression profile
- Cell type identification
Reads
RNA-Seq:
Sample RNA -> Amplified cDNA -> cDNA fragments -> reads
A short DNA fragment which is read out by sequencer.
FASTQ format
Transcript abundance in RNA-seq
RPKM:the number of mapped Reads per KB per million reads.
Mapping Reads from RNA-Seq
Detection novel splicing isoforms through junction reads
Handle Junction Reads:
“Join exon” strategy
- Steps
- Build “conceptual junctions library” for each known transcript
- Map RNA-Seq reads to the genome and CJL
- Fast
- Can identify novel splicing isoforms
- Can NOT find novel exons and novel genes
- Steps
"Split reads" strategy
- Steps
- Unsplicingly map to genome
- For failed mapped reads, split them into several k-mer seeds
- Stitch mapped seeds together as whole read alignment
- Slower
- Can identify novel splicing isoforms and find novel exons and novel genes
- Steps
Genome assembly(组装)
Overlap - Layout - Consensus (OLC): well established,more powerful method, but more difficult to implement. First to be used successfully for complex Eucaryotic genomes(Drosophila,H.sapiens)
DeBruijn - Euler: newer, easier to implement, problematic in complex genomes (for current implementations)
OLC Steps:
- Find Overlaps by aligning the sequence of the reads
- Layout the reads based on which aligns to which
- Get Consensus by joining all read sequences, merging overlaps
- Sequencer reads in random direction, left-to-right or right-to-left
- Change into Shortest Common Superstring (SCS) -> Hamiltonian Cycle or Traveling Saleman Problem
DeBruijn Steps:
- The de Bruijn graph was developed outside the realm of DNA sequencing to represent strings from a finite alphabet.
- The nodes represent all possible fixed-length strings.
- The edges represent suffix-to-prefix perfect overlaps.
- A K-mer graph is a form of de Bruiin graph. Its nodes represent all the fixed-length subsequences (k-mers) drawn from a read. Its edges represent all the fixed-length overlaps between subsequences.
- Then find the Eulerian path
Sequence Alignment in Biology
The purpose of a sequence alignment is to line up all residues in the inputted sequence(s) for maximal level of similarity, in the sense of their functional or evolutionary relationship.
Pairwise Sequence Alignment
Obviously, we have(global):
for loacal:
BLAST
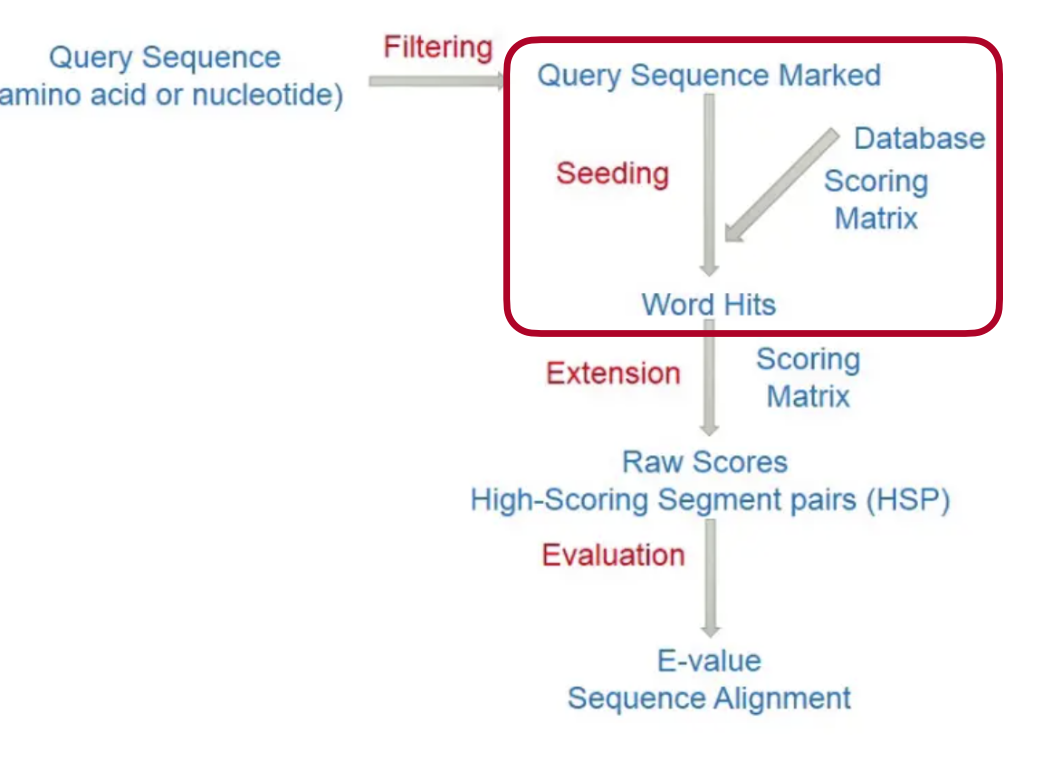
Seeding: - Hash - Burrows-Wheeler transform
Extension: - Hit cluster - Smith-Waterman algorithm(local alignment)
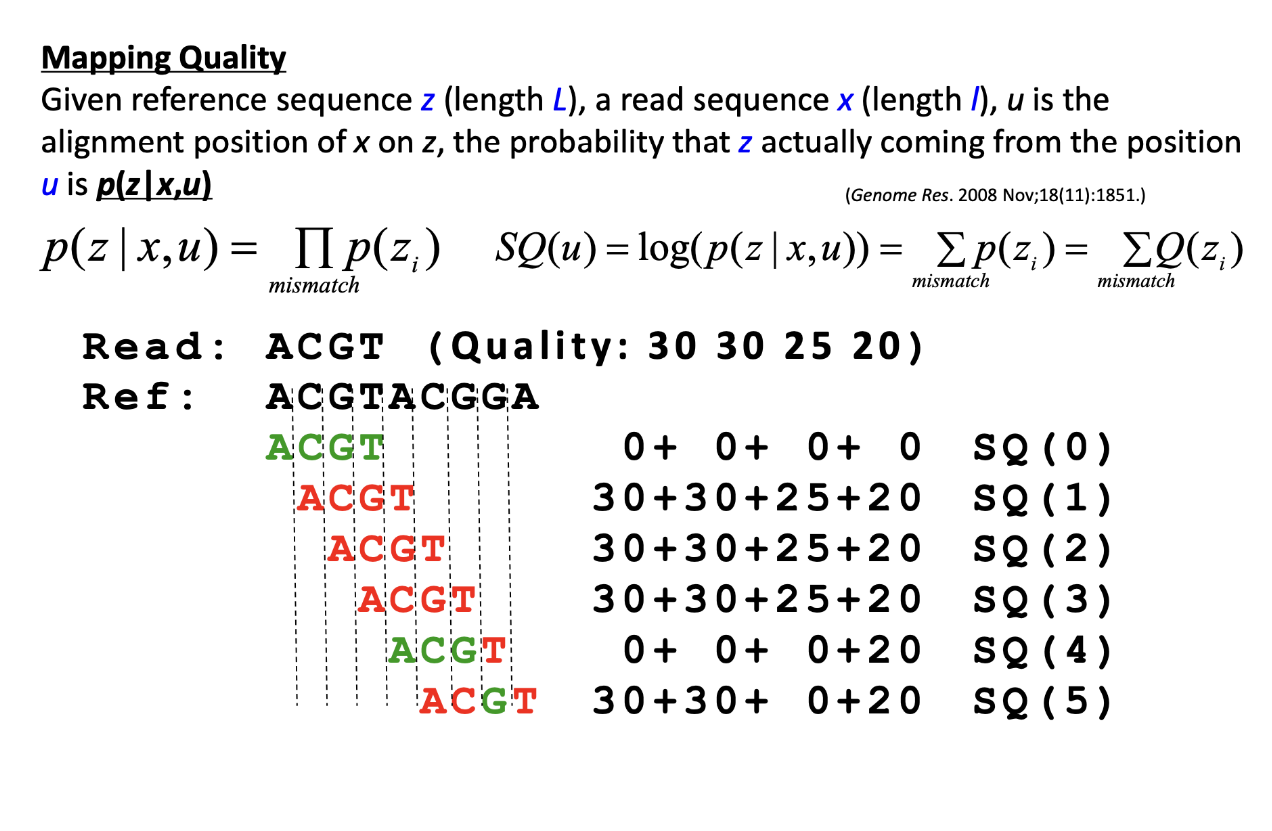
Mapping Quality:
Signature
a protein category such as a domain or motif
Domain
- A region of a protein that can adopt a 3D structure
- A fold
A family is a group of proteins that share a domain
Motif (or fingerprint):
- a short, conserved region of a protein
- typically 10 to 20 contiguous amino acid residues
CRISPR-Cas systems
The CRISPR-Cas system in bacteria serves as a defense mechanism against foreign genetic material, such as viruses and plasmids.
Cas nucleases induce DNA double-strand breaks (DSB) at desired locations within a genome.
DSBs are themselves highlygenotoxic lesions and as such cells have evolved multiple mechanisms for their repair
- NEHJ: 快速修复
- HDR: 精确修复
CRISPR-Cas systems can use as a Genome editing tools,using Artificially designed and synthesized Cas9 expression vector:
- In silico target/gRNA design
- Expression vector construction
- Expression vector delivery
CRISPR-Cas system is aprogrammable genome editing tools
Chapter3 Data and Resource
Gene Ontology
- Molecular Function = elemental activity/task carbohydrate binding and ATPase activity
- Biological Process = biological goal or objective
- Cellular Component= location or complex
Relation:
- is a
- part of
- regulate(调节)
- pos/neg
Sequence formats: FASTA
Alignment formats: SAM (Sequence Alignment Map)
Features/annotations formats
- VCF (Variant Calling Format/File)
- GFF (General Feature Format or Gene Finding Format)
- PDB (Protein Data Bank formats)

